

原文链接:https://doi.org/10.1093/nar/gkaa920
参考文献
2.Ruddigkeit, L., van Deursen, R., Blum, L.C. and Reymond, J.L. (2012) Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J. Chem. Inf. Model., 52, 2864-2875.
3.Levré, D., Arcisto, C., Mercalli, V. and Massarotti, A. (2019) ZINClick v.18: Expanding Chemical Space of 1,2,3-Triazoles. J. Chem. Inf. Model., 59, 1697-1702.
4.Segler, M.H.S., Kogej, T., Tyrchan, C. and Waller, M.P. (2018) Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent Sci, 4, 120-131.
5.Zhavoronkov, A., Ivanenkov, Y.A., Aliper, A., Veselov, M.S., Aladinskiy, V.A., Aladinskaya, A.V., Terentiev, V.A., Polykovskiy, D.A., Kuznetsov, M.D., Asadulaev, A. et al. (2019) Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol., 37, 1038-1040.
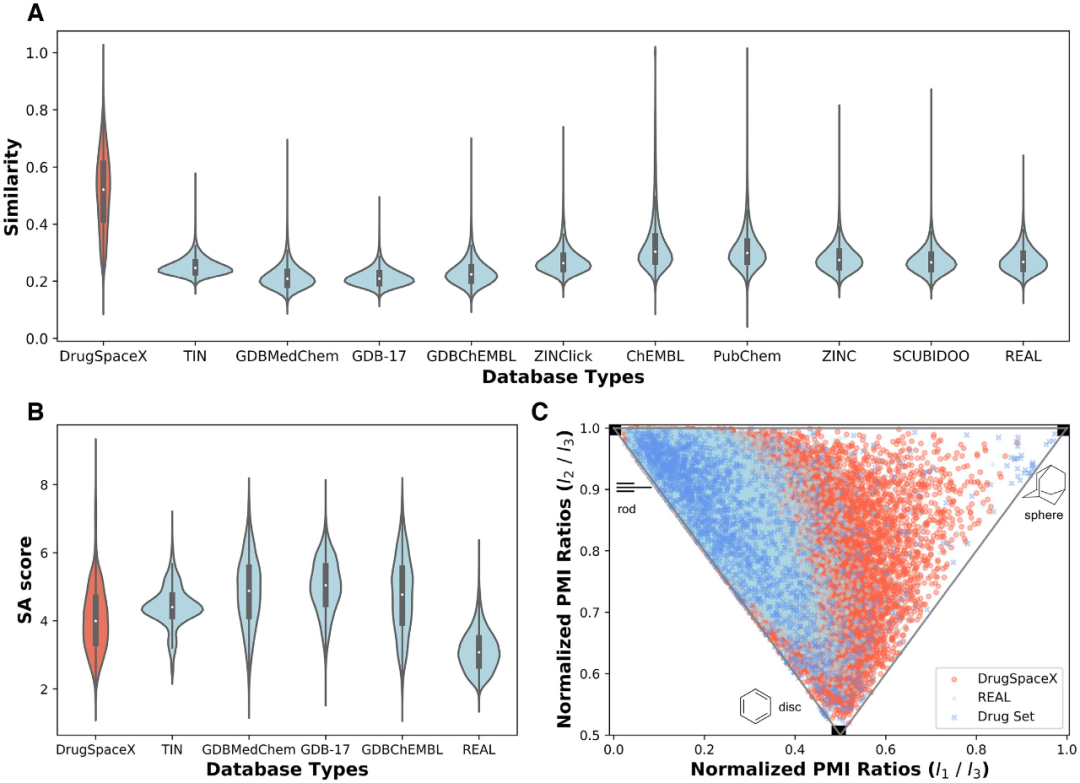
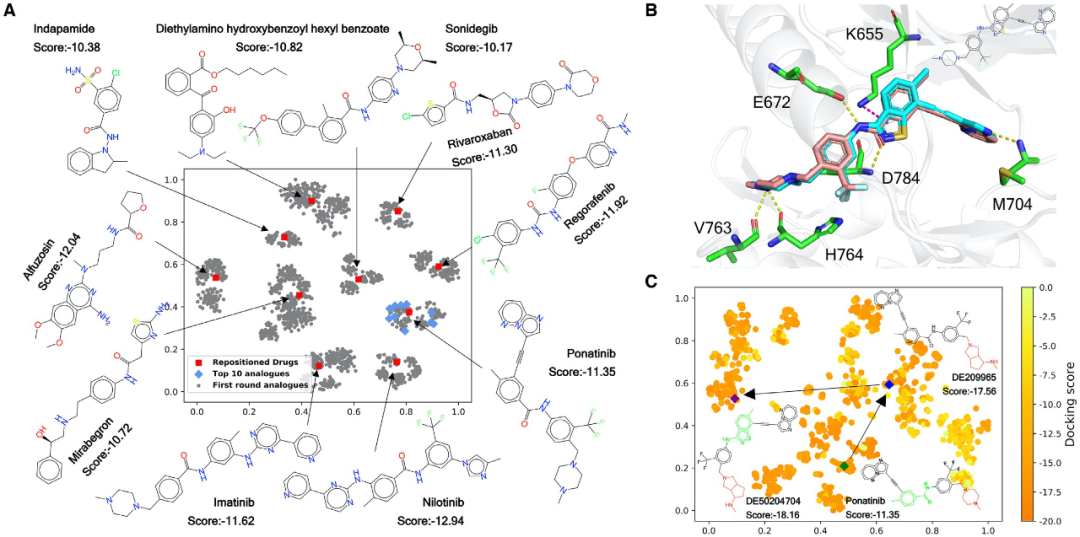
6.Yang, T., Li, Z., Chen, Y., Feng, D., Wang, G., Fu, Z., Ding, X., Tan, X., Zhao, J., Luo, X. et al. (2020) DrugSpaceX: a large screenable and synthetically tractable database extending drug space. Nucleic Acids Research.
7.Gorgulla, C., Boeszoermenyi, A., Wang, Z.F., Fischer, P.D., Coote, P.W., Padmanabha Das, K.M., Malets, Y.S., Radchenko, D.S., Moroz, Y.S., Scott, D.A. et al. (2020) An open-source drug discovery platform enables ultra-large virtual screens. Nature, 580, 663-668.
中国药学Cell丨着眼破解EZH2抑制剂临床应用的困局——张毅、蒋华良点评
总生存率增长141%!维持化疗前测下这个免疫治疗,可判断治疗效果
蛋白靶向降解:除了PROTAC,还有LYTAC、AUTAC和ATTEC

 点击直达,每月2万多朋友到过这里!
点击直达,每月2万多朋友到过这里!
发布者:药时代,转载请首先联系contact@drugtimes.cn获得授权

 为好文打赏 支持药时代 共创新未来!
为好文打赏 支持药时代 共创新未来! 
